博文
[PNAS, 11/2/2020]乙烯响应下TREE1及EIN3合作介导的转录抑制活动会阻碍植株地上部分的生长发育
||
TREE1-EIN3–mediated transcriptional repression inhibits shoot growth in response to ethylene
Author: Likai Wang, et al.
Corresponding author: Hong Qiao.
PNAS first published November 2, 2020
Affiliations:Institute for Cellular and Molecular Biology, The University of Texas at Austin, Austin, TX 78712; and bDepartment of Molecular Biosciences, The University of Texas at Austin, Austin, TX 78712
时隔多日的文献精读~这段时间想多看一些转录因子的文章,这篇文章会作为一门课程的汇报来讲,希望自己加油~
【ABSTRACT】
Ethylene is an important plant hormone that regulates plant growth, in which the master transcriptionactivator EIN3 (Ethylene Insensitive 3)-mediated transcriptional activation plays vital roles. However, the EIN3-mediated transcriptional repressionin ethylene response is unknown. We report here that a Transcriptional Repressor of EIN3-dependent Ethylene-response 1 (TREE1) interacts with EIN3 to regulate transcriptional repression that leads to an inhibition of shoot growth in response to ethylene. Tissue-specific transcriptome analysis showed that most of the genes are downregulated by ethylene in shoots, and a DNA binding motif was identified that is important for this transcriptional repression. TREE1 binds to the DNA motif to repress gene expression in an EIN3-dependent manner. Genetic validation demonstrated that repression of TREE1-targeted genes leads to an inhibition of shoot growth. Overall, this work establishes a mechanism by which transcriptional repressor TREE1 interacts with EIN3 to inhibit shoot growth via transcriptional repression in response to ethylene.
乙烯是植物中非常重要的一类调控生长发育的激素,EIN3(Ethylene Insensitive 3,乙烯不敏3 )是其中非常重要的转录激活因子。然而,EIN3介导的乙烯信号转录活动中的抑制过程依旧没有研究清楚。本文报道了一个转录抑制因子TREE1(Transcriptional Repressor of EIN3-dependent Ethylene-response 1,乙烯响应EIN3依赖型转录抑制因子1)与EIN3互作来调控转录抑制,导致植株对乙烯响应后地上部分的生长被抑制。组织特异转录组分析结果显示,地上部分中多数基因在乙烯处理后表达下调,并且我们还发现其中有一个DNA结合基序(DNA binding motif)在这个转录抑制活动中起到重要作用。TREE1与该DNA结合基序结合从而在EIN3依赖活动中下调基因表达。基因验证(genetic validation)表明,TREE1的靶向基因的表达抑制导致了植株地上部分生长发育的抑制。总而言之,本文的工作揭示了这样一个机制——乙烯处理下,转录抑制因子因子TREE1与EIN3互作介导转录抑制导致植株地上部分发育抑制。
【INTRODUCTION】
Ethylene is one of the most important plant hormones that is essential for many physiological and developmental processes such as apical hook formation, shoot and root growth, root nodulation, flower senescence, abscission, and fruit ripening (1). Plants also produce ethylene to fight against wounding, pathogen attack, or stress threats such as extreme temperatures or drought (2–4). Ethylene is perceived by a family of receptors that bind to the endoplasmic reticulum (ER) membrane (5–8). In the absence of ethylene, the receptor ETR1 (ETHYLENE RESPONSE 1) interacts with CTR1 (CONSTITUTIVE TRIPLE RESPONSE 1), a Raf-like protein that phosphorylates EIN2 (ETHYLENE INSENSITIVE 2), the key positive regulator of ethylene signaling, preventing the ethylene response(5,9–13). In the presence of ethylene, both ethylene receptors and CTR1 are inactivated; the C terminus of EIN2 is dephosphorylated and cleaved by unknown mechanisms. The cleaved C-terminal end of EIN2 is translocated into the nucleus (14–16), where it interacts with a histone binding protein ENAP1 (EIN2 NUCLEAR ASSOCIATED PROTEIN 1) to regulate the histone acetylation at H3K14 and H3K23 to integrate transcriptional regulation that is mediated by transcription factors EIN3 (ETHYLENE-INSENSITIVE 3) and EIL1 (ETHYLENE-INSENSITIVE3-LIKE 1) (17).
在植株的各种生理与发育过程中,包括顶端弯钩(apical hook)的形成,地上与地下部分的生长,根瘤形成(root nodulation),花器官老化及脱落(flower senescence & abscission),以及果实成熟(ripening),乙烯都是非常重要的植物激素[1]。乙烯也常常被植物用于抵抗伤口(wounding),病毒入侵(pathogen attack)以及逆境胁迫(如极端温度以及干旱)[2-4]。当乙烯缺失时,受体ETR1(ETHYLENE RESPONSE 1)与CTR1(CONSTITUTIVE TRIPLE RESPONSE 1)互作来保证乙烯响应,其中CTR1是一种类Raf蛋白,可以磷酸化EIN2(ETHYLENE INSENSITIVE 2),EIN2正是乙烯响应的关键积极调控因子[5,9-13]。当乙烯存在时,CTR1与受体(如ETR1)都会处于非活性状态,而EIN2的羧基端(C terminus)会被脱磷酸化,并被切割开——这一过程的机制至今仍不清楚。EIN2被切割的羧基端会转移到核中[14-16],在这里它与一个组蛋白结合蛋白ENAP1(EIN2 NUCLEAR ASSOCIATED PROTEIN 1)结合,调控H3K14与H3K23的组蛋白乙酰化,协助完成转录因子EIN3(ETHYLENE-INSENSITIVE 3)与EIL1(ETHYLENE-INSENSITIVE3-LIKE 1)的转录调控过程[17]。
博主注:幼苗顶端弯钩的形成是双子叶植物暗形态建成过程中的一个重要事件。它保护双子叶植物幼嫩的子叶和脆弱的顶端分生组织在幼苗出土时免受机械损伤,进而保证幼苗的出苗率和成活率。幼苗顶端弯钩的形成是由于下胚轴顶端的两个对立面之间细胞分裂和延伸的不对称所引起的。乙烯属于一类与内质网(endoplasmic reticulum, ER)膜结合的受体因子家族[5-8]。
The typical dynamic physiological response to the presence of ethylene is a rapid growth inhibition that depends on the master transcriptional regulator EIN3 (18). Both genetic and molecular studies have demonstrated that EIN3 and EIL1 are the positive regulators that are necessary and sufficient for the ethylene response (19–21). By analyzing the promoters of the genes that are highly up-regulated by ethylene, the EIN3 binding motif was identified and then validated by using an electrophoresis mobility shift assay (EMSA) (22–28). The binding motif was further verified by chromatin immunoprecipitation sequencing (ChIP-seq) of EIN3 over a time course of ethylene treatments and following data analysis (21). All of this research showed that EIN3 is a transcription activator and plays a key role in the ethylene response. Whereas the time course transcriptome analysis in response to ethylene revealed that most of the genes are repressed by ethylene in the early stage (before 1 h) of the ethylene response, about 50% of ethylene-altered genes are repressed in the later stage (after 1 h) of ethylene response (18, 21). More interestingly, parts of the ethylene-repressed genes are the binding targets of EIN3, the transcription activator (21), suggesting that EIN3 is potentially involved in the transcriptional repression. However, little research has been focused on the EIN3-mediated transcriptional repression in the ethylene response. We recently showed that histone deacetylases SRT1 (SIRTUIN 1) and SRT2 (SIRTUIN 2) are involved in the transcriptional repression in the ethylene response by maintaining a low level of H3K9Ac in the promoters of ethylene-repressed genes (29). Yet, how the EIN3-mediated transcriptional repression occurs and whether other factors are involved are still unknown.
对于乙烯存在的典型动态生理响应(typical dynamic physiological response)就是迅速的生长抑制——依赖于主要转录调控因子EIN3[18]。无论是基因或者分子层面的研究都表明EIN3与EIL1都是在乙烯响应过程中非常必要且关键的转录激活因子[19-21]。通过对乙烯处理下表达高度上调的基因的启动子的分析发现了一种EIN3结合的基序,后续通过对EMSA实验也证明了这个结论[22-28]。另外对EIN3的CHIP-seq(chromatin immunoprecipitation sequencing)实验分析也进一步证实了这个结合基序的存在[21]。所有的这些研究结果表明,EIN3是一种转录激活子,并且在乙烯响应过程中发挥关键作用。然而在对乙烯处理的转录组进行时间分析爱后发现,其实在对乙烯响应的前期(1h内)大多数基因都是被抑制表达的,而在1h后仍旧有50%的基因被抑制表达[18,21]。更有趣的是,一部分乙烯抑制表达的基因甚至是转录激活子EIN3的靶向结合基因[21],这表明EIN3可能还参与了一些转录抑制活动。然而,对于EIN3介导的乙烯响应下的转录抑制活动的研究寥寥无几。我们最近的研究表明组蛋白去乙酰酶SRT1以及SRT2通过维持乙烯抑制基因的启动子中H3K9Ac的低水平含量,来参与乙烯响应下的转录抑制[29]。然而,EIN3介导的转录抑制的发生机制以及是否有其它组分因子参与其中,至今未知。
In this study, by analyzing tissue-specific ethylene-regulated gene expression, we found that most of the genes are downregulated by ethylene in shoots. By a motif search in the promoters of the ethylene-repressed genes in shoots, we identified a DNA binding motif that can be recognized by DUO1-ACTIVATED ZINC FINGER (DAZ) proteins. Further, we found that a Transcriptional Repressor of EIN3-dependent Ethylene-response 1 (TREE1) binds to the specific DNA motif to repress gene expression by both ChIP-seq and ChIP-qPCR. Importantly, TREE1 interacts with EIN3 that is required for TREE1 binding and transcriptional repression regulation on its targets in response to ethylene in shoots. Genetic validation demonstrated that the transcriptional repression of TREE1-targeted genes leads to an inhibition of shoot growth in response to ethylene. Overall, this work establishes a mechanism by which transcriptional repressor TREE1 interacts with EIN3 to inhibit shoot growth via transcriptional repression regulation in response to ethylene.
在本次研究中,通过对组织特异的乙烯调控的基因的表达分析,我们发现植株地上部分(以下简称shoot)的绝大多数的乙烯调控的基因表达都下调了。通过对shoot中乙烯抑制表达基因的启动子基序查询,发现了一种可以被DAZ(DUO1-ACTIVATED ZINC FINGER,DUO1激活型锌指蛋白)蛋白识别的DNA结合基序。另外,通过CHIP-seq以及CHIP-qPCR我们还发现一个乙烯响应下EIN3依赖型的转录抑制子TREE1可以与特异性DNA基序结合来抑制基因表达。更重要的是,shoot中乙烯响应下,TREE1与DNA基序的结合以及对下游目的基因的转录抑制的调控,都需要TREE1与EIN3互作。基因验证表明乙烯响应下,TREE1对下游目的基因的转录抑制对导致shoot生长的抑制。总之,本文的工作揭示了这样一个机制——乙烯处理下,转录抑制因子因子TREE1与EIN3互作介导转录抑制导致植株地上部分发育抑制。
【RESULTS】
1.A Unique DNA Binding Motif Is Present in the Promoters of Down-Regulated Genes by Ethylene in Shoots.
1.shoot中乙烯响应下表达下调的基因启动子中存在一个独特的DNA结合基序
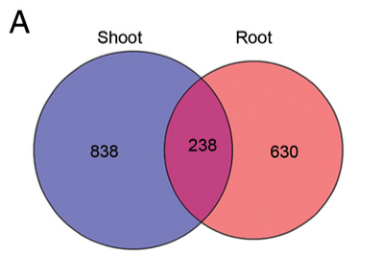
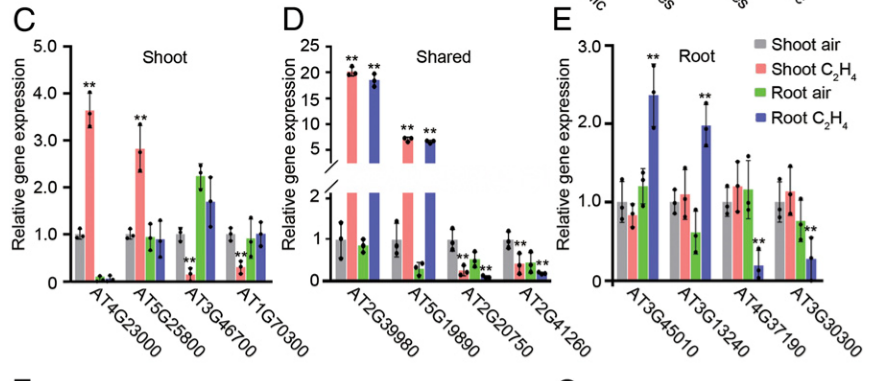
To explore how different tissues respond to ethylene, we reanalyzed previously published RNA-seq data collected from the shoots and the roots of 3-d-old etiolated Arabidopsis Col-0 seedlings treated with air or 4 h of ethylene (30). In this paper, the shoots indicate the tissues other than roots, including the seedling hypocotyls and apical hooks. In total, 1,076 differentially expressed (DE) genes by ethylene treatment were identified from shoots and 868 DE genes were identified from roots (Fig. 1A). Among them, only 238 genes were differentially regulated in both tissues; 838 genes were specifically regulated by ethylene in shoots and 630 genes were specifically regulated in roots (Fig. 1A).
为了探究不同的组织如何对乙烯进行响应,我们重新分析了之前曾经发表过的转录组测序数据(Col型拟南芥幼苗3天黄化幼苗的shoot与root部分,分别置于CK或4h乙烯处理)[30]。在这篇文章中,shoots指的是除了地下组织(root)以外的组织,包括幼苗下胚轴(hypocotyls)以及顶端弯钩(apical hook)。总体而言,乙烯处理后,shoot中共有1076个基因出现了差异表达(differentially expressed, DE)而roots中则有868个(Fig. 1A)。在它们之间,只有238个基因在两种组织中(指root与shoot中)共同差异表达;即838个基因只在shoot中表达,630个基因只在root中表达(Fig. 1A)。
博主注:A gene is declared differentially expressed if an observed difference or change in read counts or expression levels between two experimental conditions is statistically significant.
By further comparing the RNA-seq datasets, we found that about 60% of ethylene-regulated DE genes from 3-d-old etiolated whole seedlings were different from tissue-specific ethylene-regulated DE genes (SI Appendix, Fig. S1A). Gene Ontology (GO) analysis showed that ethylene-related GO terms were enriched only in the DE genes identified both in shoots and in roots (SI Appendix, Fig. S1B). Root development-related GO terms were specifically enriched in the DE genes in roots (SI Appendix, Fig. S1C). Stress response genes were specifically enriched in the DE genes in shoots (SI Appendix, Fig. S1D).
对转录组分析数据进一步研究后发现,大约60%的乙烯调控差异表达基因(指整个seedling)与组织特异(指shoot或root)乙烯调控差异表达基因不同(Fig. S1A)。GO(Gene Ontology, 基因本体论)分析表明,乙烯相关的GO术语只在seedling中共同差异表达基因中富集(Fig. S1B)。与root发育相关的GO术语只在root差异表达基因中富集(Fig. S1C)。而胁迫响应基因则只在shoot差异表达基因中富集(Fig. S1D)。
博主注:Gene Ontology (GO) term enrichment(基因本体论术语富集)
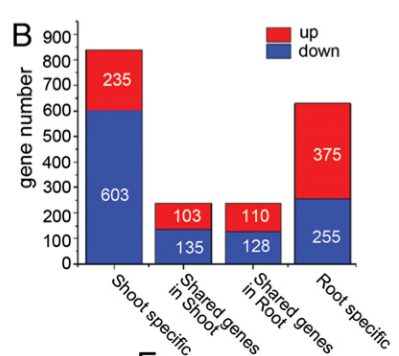
We further found that about 40% of root-specific DE genes were down-regulated, and about 50% of the shared DE genes between shoots and roots were down-regulated (Fig. 1B). Whereas more than 70% of shoot-specific DE genes were down-regulated, many of these genes are involved in the stress responses (Fig. 1B). qRT-PCR evaluation and the Integrative Genomics Viewer (IGV) data further confirmed that transcriptional patterns in different tissues are tissues distinct (Fig. 1 C–E and SI Appendix, Fig. S1 E–H).
我们继续发现大约40%的只在root中差异表达的基因(以下简称DE)表达下调了,大约50%shoot与root中共同DE基因表达下调(Fig. 1B)。然而只在shoot中的DE基因有70%都下调表达了,而这其中非常多的基因都与胁迫响应有关(Fig. 1B)。qRT-PCR以及Integrative Genomics Viewer (IGV,综合基因组查看器) 结果也进一步证明不同组织中的转录模式是不同的(Fig. 1 C–E and SI Appendix, Fig. S1 E–H)。
其中ACTIN2 gene expression作为参考来体现基因表达量(前两个上调,后两个下调),**代表与air条件的显著差异。Fig.S1 F-H为Genome Browser annotation tracks(基因组浏览器注释轨道,体现转录组分析数据)。
To investigate why more down-regulated genes by ethylene treatment appeared in shoots, we conducted motif searches in the promoter regions of ethylene up-regulated genes and ethylene down-regulated genes in both tissues. Only the EIN3 binding motif was identified from up-regulated genes, and no other significant DNA binding motifs were identified from the downregulated genes. We then decided to narrow down the genes for motif search. Given the genes are regulated by ethylene and EIN3 is the key transcription factor that regulates gene expression in response to ethylene, we decided to identify EIN3 target genes for further analysis.
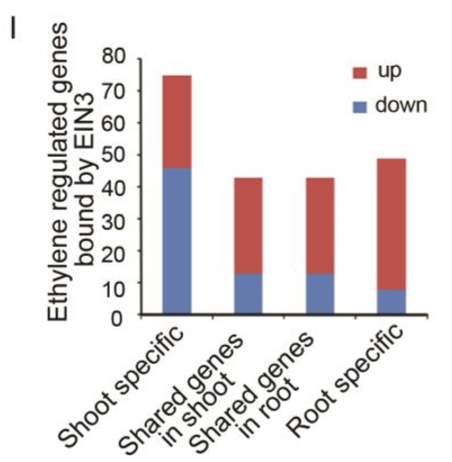
By comparing the ethylene-regulated genes in different tissues with the EIN3 binding target genes (21), we identified 75, 49, and 43 EIN3-bound genes that are regulated by ethylene in shoots, roots, and in both of the two tissues, respectively (SI Appendix, Fig. S1I). Most EIN3 targets in roots or in both tissues were up-regulated by ethylene, which is in agreement with a previous finding that the expression of EIN3 bound genes was generally enhanced by ethylene. In shoots, however, more EIN3 targets were down-regulated than upregulated (SI Appendix, Fig. S1I). ChIP-seq profile analysis revealed that the mean EIN3 binding signal in the promoters of ethylene-activated genes was significantly greater than that in ethylene-repressed genes in roots, or in the shared genes between shoot and root (SI Appendix, Fig. S1 J and K; EIN3 binding signal in shared DE genes, up vs. down: P = 9.98E-07; in root-specific DE genes, up vs. down: P = 9.75E-04). In shoots, however, the mean EIN3 binding signal intensity between ethylene-activated and ethylene-repressed genes had no significant difference (SI Appendix, Fig. S1L, P = 0.243).
为了探究为什么shoot中的乙烯响应下表达下调的基因数量比root中多,我们使用了基序检测(motif search),检测了两种组织中所有乙烯响应上调/下调基因的启动子。结果显示,只有上调基因启动子中检测出了EIN3结合基序,而下调基因中没有检测出任何显著的DNA结合基序。随后我们决定进一步精确检测的基因范围。因为所有基因都是乙烯响应调控表达的,而EIN3是乙烯响应下关键的转录调控因子,所以我们决定只精准检测分析EIN3的靶标基因。
通过对乙烯调控基因以及EIN3结合靶向基因[21]的比较,我们发现在shoot,root以及seedling(即shoot与root共同)中的乙烯调控基因中,分别有75,49以及43个EIN3结合靶向基因(Fig. S1I)。绝大多数在root以及seedling中发现的EIN3结合靶向基因都是上调表达的,这也进一步证实了之前的研究结论:大多数EIN3结合靶向基因都会被乙烯调控增强表达。然而在shoot中,EIN3结合靶向基因中上调表达的比下调的更多(Fig. S1I)。CHIP-seq分析结果显示,在root或seedling(shared DE)中,乙烯激活型基因启动子中的EIN3结合信号比乙烯抑制型要多(Fig. S1 J and K);然而在shoot中,乙烯激活基因与抑制基因之间,EIN3结合信号的强度并没有显著区别(Fig. S1L)。
(J-L)EIN3 prefers bind to the ethylene-up regulated genes that shared between (J) shoots and roots or (K) in roots, but not (L) in shoots.
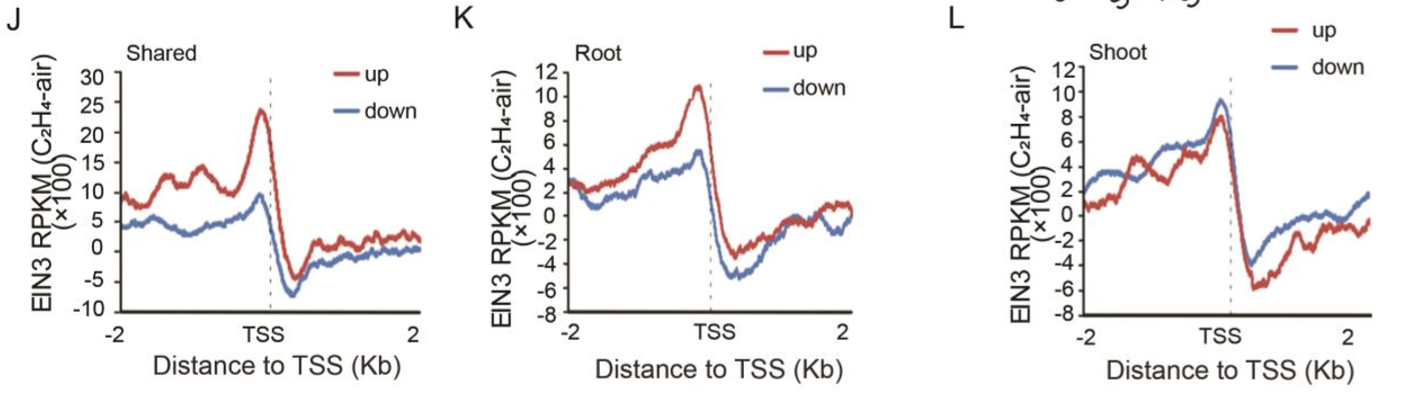
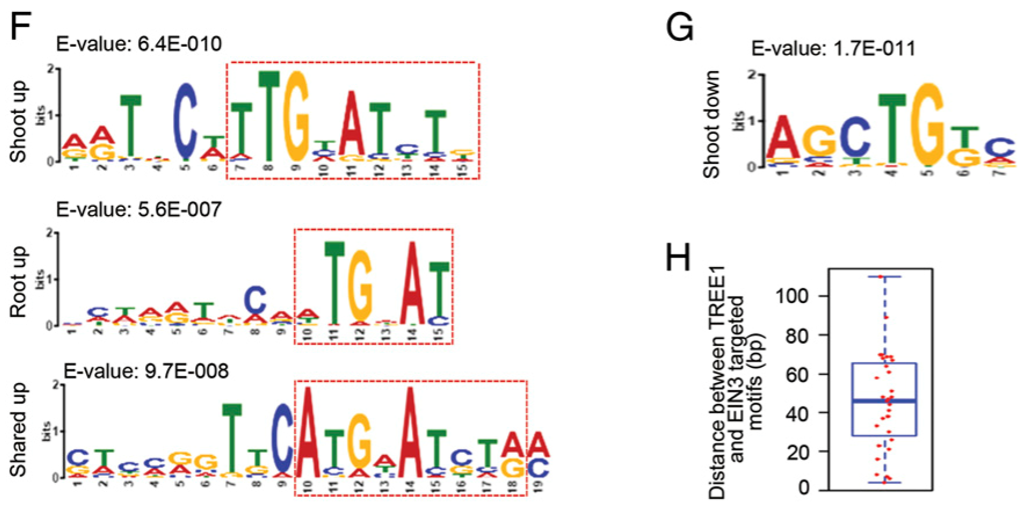
We then searched for DNA binding motifs in 200-base pair sequences centered on the EIN3 binding summit in EIN3-bound ethylene up- or down-regulated genes in shoots and in roots. The known EIN3 binding motif was identified in genes that are activated by ethylene in shoots, in roots, or in both tissues (Fig.1F). Only one DNA binding motif with the consensus sequence AGCTGT/GC/A (E value = 1.7E-011) was identified in the genes that are repressed by ethylene in shoots (Fig. 1G), while not in roots. The motif was different from the EIN3 binding motifs that were determined and tested previously (21, 28), and the distance of the motif is close to the EIN3 binding motif (Fig. 1H).
随后我们在shoot与root中的所有乙烯调控基因的(其中EIN3结合的)EIN3结合的峰值周围200bp(200-base pair)序列中,寻找新的DNA结合基序。在所有组织中,乙烯激活基因的启动子中发现了很多已知的EIN3结合基序(Fig.1F),但是乙烯抑制基因中,只有一个在shoot中的基因被发现存在一个含有共同序列AGCTGT/GC/A的DNA结合基序,但root中并没有(Fig. 1G)这个基序与之前报道过的EIN3结合基序并不相同[21,28],但与EIN3结合基序距离很近(Fig. 1H)。
PS.Motifs were found by the MEME-ChIP (meme-suite.org/tools/meme-chip).
2.The DNA Binding Motif Identified Is Involved in the Transcriptional Repression in the Ethylene Response.
2.被识别的新DNA结合基序参与了乙烯响应下的转录抑制
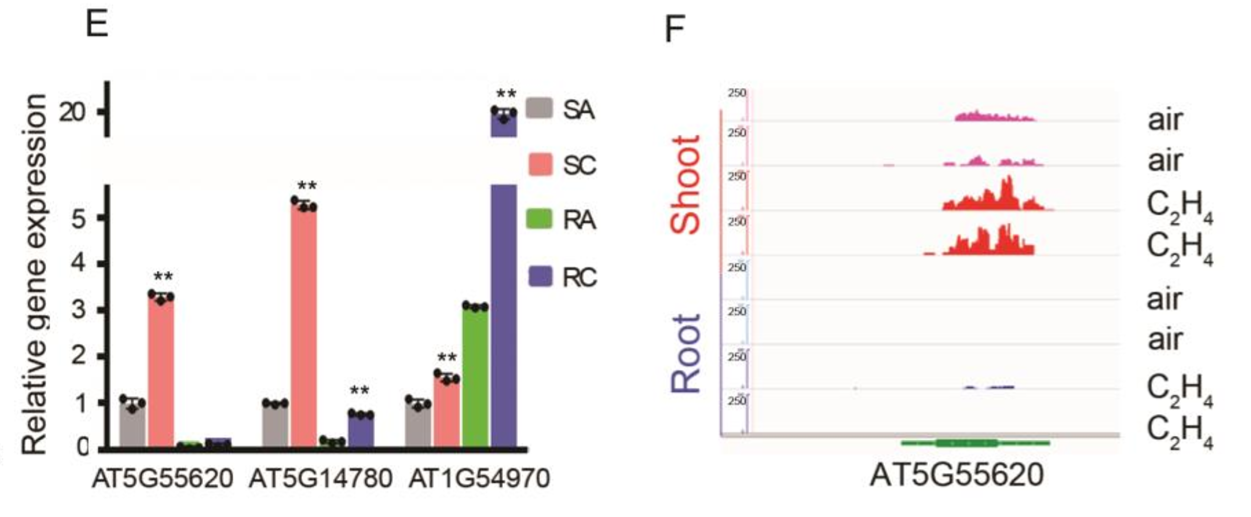
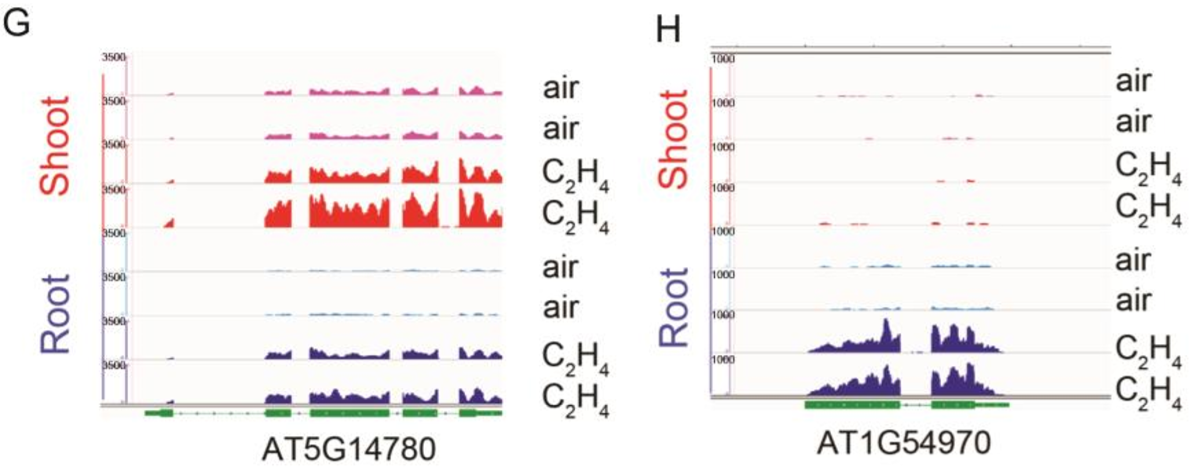
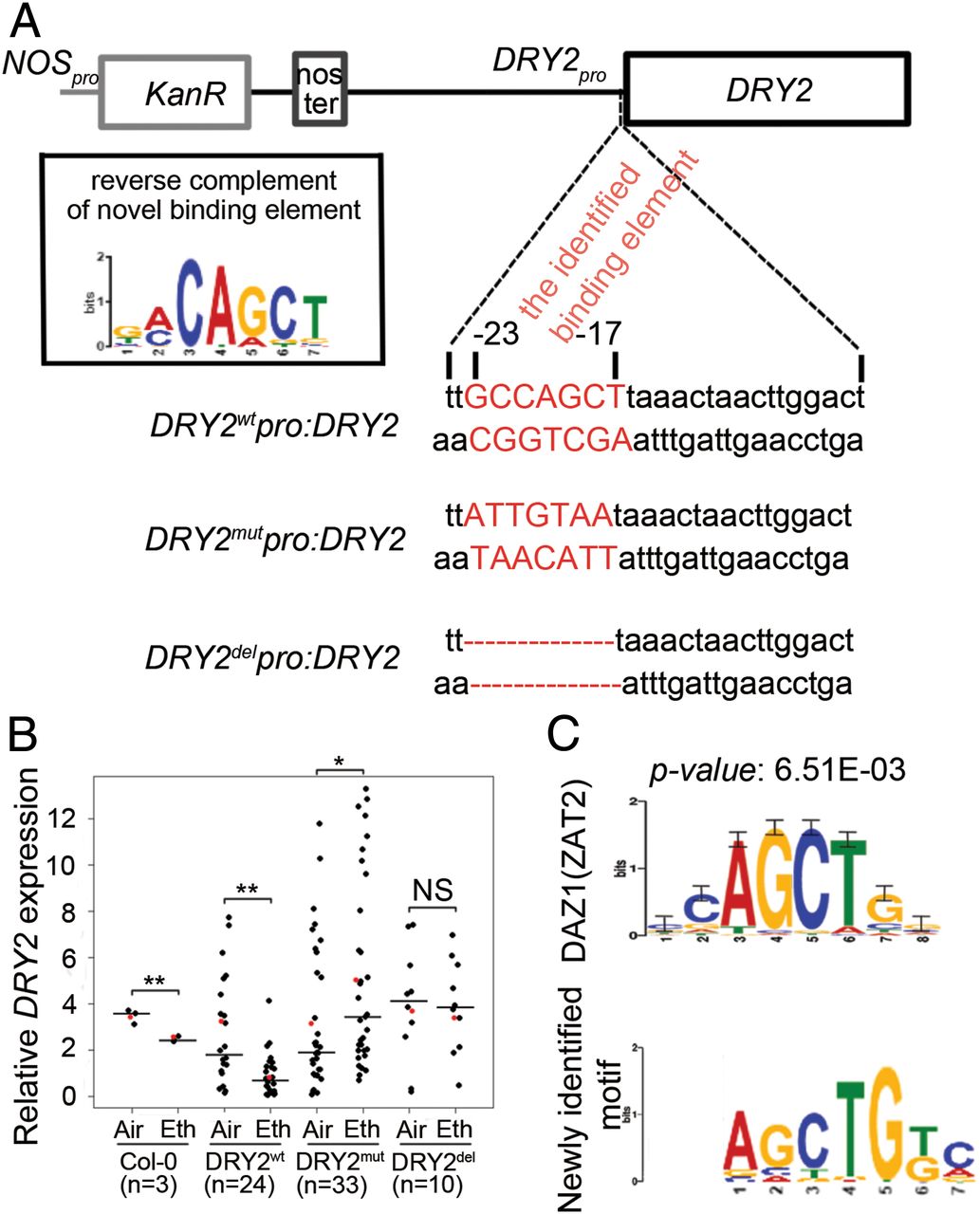
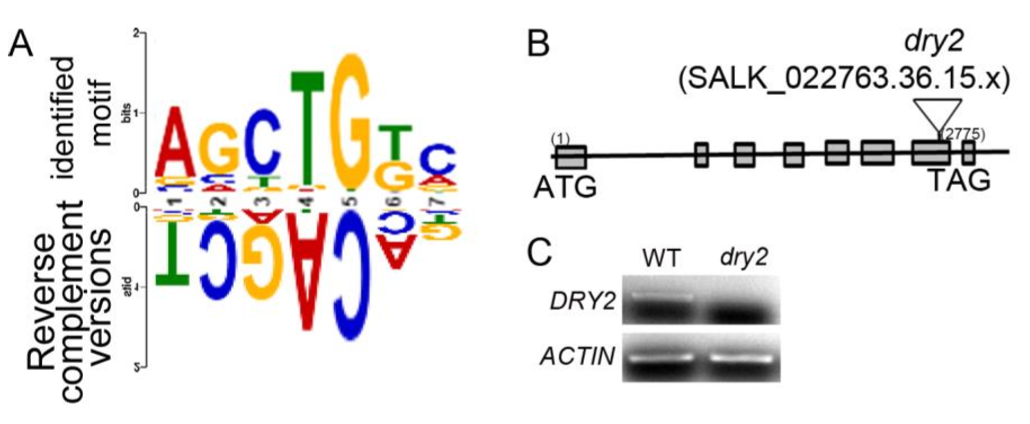
To evaluate the role of the DNA binding motif in the regulation of gene expression in response to ethylene in vivo, we identified a gene (DROUGHT HYPERSENSITIVE2;DRY2;AT1G58440)that is down-regulated by ethylene in shoots, and its promoter region contains the DNA binding motif (Fig. 2A and SI Appendix, Fig. S2 A). We then constructed a reporter gene driven by the native promoter (pDRY2wt: DRY2), by the promoter with mutations in the DNA binding motif (pDRYmut: DRY2), or by the promoter with a deletion of the DNA binding motif (pDRY2del: DRY2) (Fig. 2A and SI Appendix, Fig. S2 A). We then introduced these constructs into the dry2 mutant (salk_022763; SI Appendix, Fig. S2 B and C). By examining DRY2 gene expression in T2 plants treated with or without ethylene, we found that DRY2 gene expression was repressed by ethylene when the gene was driven by the wild-type promoter. The expression was not altered by ethylene when the gene was driven by the promoter in which the DNA binding motif was mutated or deleted (Fig. 2B). These results strongly indicate that the DNA binding motif is involved in the transcriptional repression in response to ethylene in vivo.
为了探究乙烯响应下,该DNA结合基序究竟在基因表达调控中发挥了怎样的作用,我们选择了一个基因DRY2(DROUGHT HYPERSENSITIVE2,干旱高敏2),它在shoot中受乙烯下调表达,并且其启动子中包含该DNA结合基序(Fig. 2A, Fig. S2 A),随后我们构建了一些DRY2的报告基因,包括正常自身启动子(pDRY2wt: DRY2),突变自身启动子(pDRYmut: DRY2),以及自身启动子缺失(pDRY2del: DRY2)(Fig. 2A, Fig. S2 A)。我们将这些载体转到dry2突变体中(Fig. S2 B and C)。通过对T2代植株中DRY2基因的表达水平检测(CK或乙烯处理),我们发现乙烯响应下,pDRY2wt驱动下的DRY2基因表达被抑制。而当启动子中的DNA结合基序被突变或缺失后,DRY2的表达量与CK没有极显著差异(Fig. 2B)。这些结果强有力地证明,该DNA结合基序参与到植株体内乙烯响应下的基因转录抑制。
3.EIN3 Interacts with TREE1 and Its Homolog DAZ3 Both In Vitro and In Vivo.
3.EIN3与TREE1及其同源基因DAZ3在体内与体外均存在互作
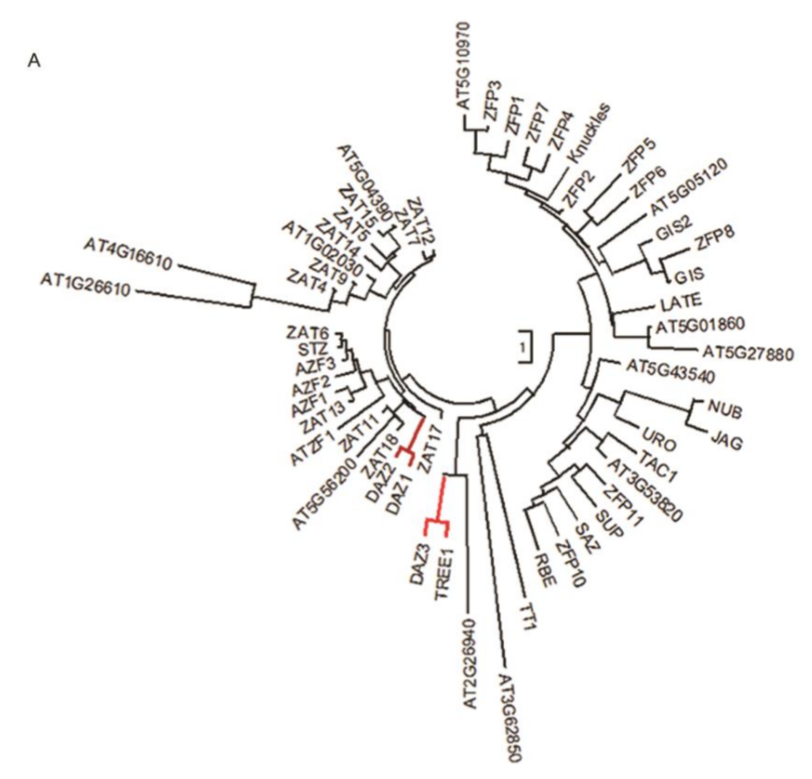
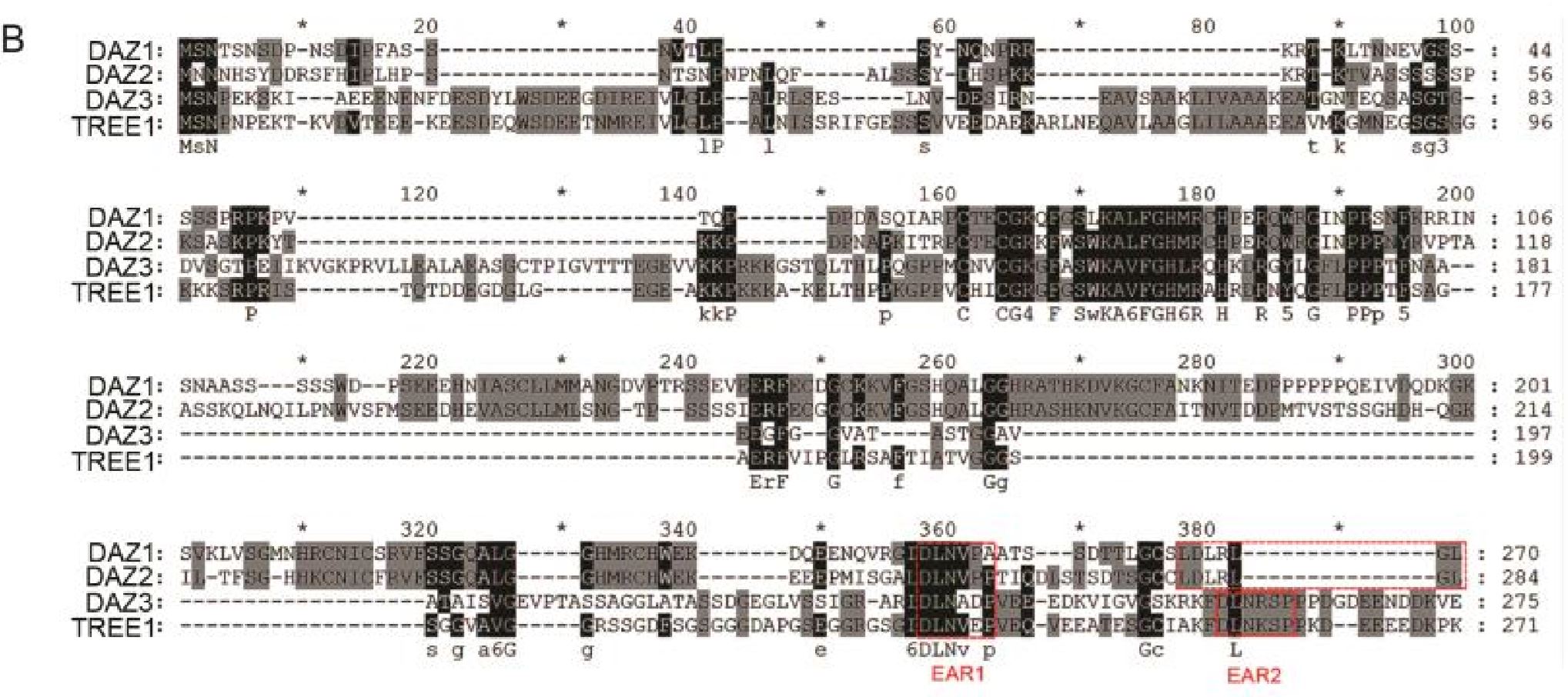
To identify the protein(s) that potentially binds to the DNA binding motif, we conducted a deep search using AGCTGT/GC/A motif to search against the database Tomtom (meme-suite.org/ tools/tomtom) (31) or literatures (32, 33). A DNA binding motif bound by transcription factor DAZ1 (also known as DUO1 ACTIVATED ZINC FINGER 1 or ZAT2) and a previously functionally uncharacterized protein AT4G35610 (Transcriptional Repressor of EIN3-dependent Ethylene-response 1, TREE1) (32, 33) showed a significant similarity to the motif we identified (Fig. 2C, E value = 7.35E-01; P value = 6.51E-03). Both DAZ1 and TREE1 belong to the C2H2 family. By phylogenetic tree analysis, we found that DAZ3 is the closest homolog of TREE1; DAZ2 is the closest homolog of DAZ1 (SI Appendix, Fig. S3A). Further analyses showed that both TREE1 and DAZs contain two ethylene-responsive element binding factor associated amphiphilic repression (EAR) motifs that are the most predominant form of transcriptional repression motif in plants (SI Appendix, Fig. S3B) (34, 35).
为了探究与可能与该DNA结合基序进行结合的蛋白,我们利用AGCTGT/GC/A基序在Tomtom数据库(meme-suite.org/ tools/tomtom)[31]以及文献[32,33]中进行了一个深入搜查。我们发现转录因子DAZ1(DUO1 ACTIVATED ZINC FINGER 1,DUO1激活型锌指蛋白1)以及之前未知功能的蛋白AT4G35610(即TREE1,Transcriptional Repressor of EIN3-dependent Ethylene-response 1)[32,33]所结合的DNA基序与之前我们所研究的基序非常相似(Fig. 2C)。DAZ1及TREE1都属于C2H2家族。通过系统发生树(phylogenetic tree)分析,我们发现DAZ3是TREE1最相近的同源基因(homolog),而DAZ2则是DAZ1的最近同源基因(Fig. S3A)。进一步研究发现,TREE1及DAZ都包含两个乙烯响应原件结合因子相关两亲抑制基序(ethylene-responsive element binding factor associated amphiphilic repression motif, EAR motif),而EAR基序正是植物中最主要的转录抑制活动中的基序(Fig. S3B)。
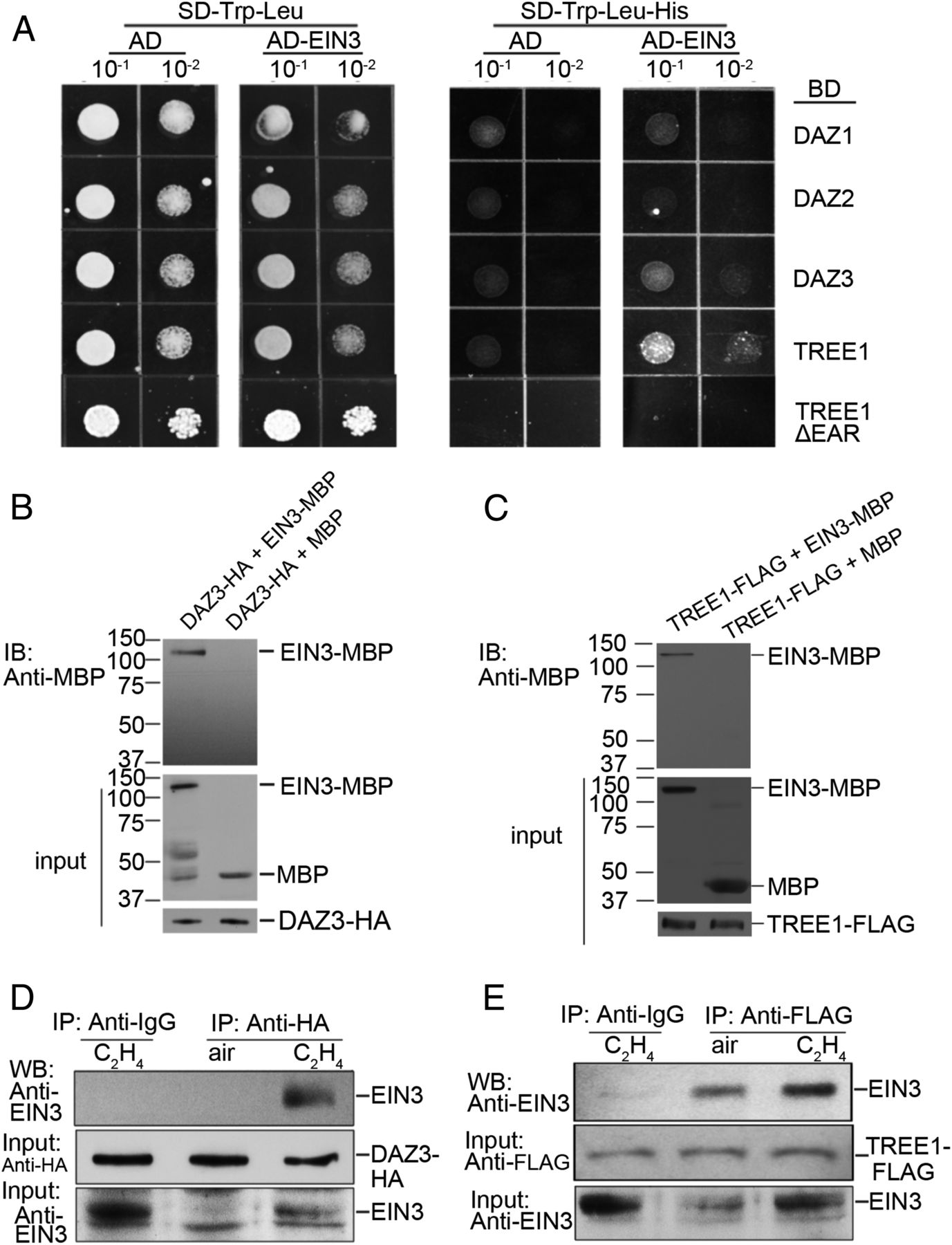
Given the motif was identified from the promoter regions of EIN3 binding targets, and it is proximate to the EIN3 binding motif, we speculated that the protein candidates can potentially interact with EIN3. Therefore, we decided to detect the physical interaction between EIN3 and TREE1 or between EIN3 and the DAZ proteins (SI Appendix, Fig. S3B) by a yeast two-hybrid assay. A strong interaction was detected between EIN3 and TREE1, and a weak interaction was detected between DAZ3 and EIN3 (Fig. 3A). But, no interaction was detected between EIN3 and DAZ1 or DAZ2 (Fig. 3A). Furthermore, we found that the deletion of EAR motifs impaired the interaction between EIN3 with TREE1 or DAZ3 (Fig.3A). We next confirmed the interaction between EIN3 and TREE1 or DAZ3 by an in vitro pull-down assay using recombinant proteins purified from Escherichia coli or Nicotiana benthamiana (Fig. 3 B and C). Finally, an immunoprecipitation assay using the extracts from TREE1-mCherry-FLAG or DAZ3-YFP-HA transgenic plants treated with or without 4 h of ethylene also demonstrated the interaction between EIN3 and TREE1 or DAZ3 in vivo, specifically in the presence of ethylene (Fig. 3 D and E). Notably, mutations in EAR motifs abolished the interaction between EIN3 and TREE1 or DAZ3 (SI Appendix, Fig. S3 C–E).
由于我们研究的基序与EIN3结合基序非常相近,我们猜测候选蛋白很可能会与EIN3互作。因此,我们决定首先运用Y2H实验去探究EIN3与TREE1或EIN3与DAZ之间是否存在互作(Fig. S3B)。结果显示,EIN3与TREE1之间存在强有力地互作,EIN3与DAZ3之间则存在微弱互作,但EIN3与DAZ1/2之间均不存在互作(Fig. 3A)。另外,我们发现当EAR基序缺失时,EIN3与TREE1及DAZ3的互作会被削弱(Fig.3A)。随后我们通过体外IP实验进一步验证了它们之间的互作,其中运用的蛋白均为从大肠杆菌或烟草中纯化(Fig. 3 B and C)。最后,我们运用TREE1-mCherry-FLAG及DAZ3-YFP-HA载体转基因植株做了体内IP实验,更进一步验证了乙烯响应下EIN3与这两者的互作(Fig. 3 D and E)。另外EAR继续的突变会导致EIN3与TREE1及DAZ3之间不再存在互作(Fig. S3 C–E)。
4.TREE1 Is a Transcriptional Repressor and EIN3 Enhances the TREE1-Mediated Transcriptional Repression.
4.TREE1是转录抑制因子,EIN3增强了TREE1介导的转录抑制活动
In order to examine the binding activity of TREE1 to TREE1 binding motif, we first conducted EMSA. Strong binding of TREE1 to TREE1 binding motif or the DRY2 promoter region that contains the TREE1 binding motif was detected, whereas no binding activity was detected when the motifs were mutated (Fig. 4 A and B and SI Appendix, Fig. S4A). We then named the binding motif as TREE1 binding motif. We next decided to evaluate the function of TREE1 in the transcriptional regulation. We conducted a transient coexpression assay in N. benthamiana using Agrobacterium that expressed a strain with luciferase driven by a minimum 35S promoter region that contained five EIN3 binding motifs and six TREE1 motifs, and a second strain for the expression of GUS, EIN3, or TREE1 (Fig. 4 C and D and SI Appendix, Fig. S4A). Compared to the control with the presence of GUS protein, the luciferase signal was significantly elevated with the presence of EIN3 (Fig. 4 E–G and SI Appendix, Fig. S4B). In contrast, the expression of luciferase was suppressed in the presence of TREE1 (Fig. 4 E–G and SI Appendix, Fig. S4B). Notably, the expression of luciferase was even lower when both TREE1 and EIN3 were present compared to when only TREE1 was expressed (Fig. 4 E–G and SI Appendix, Fig. S4B). When the TREE1 binding motif was mutated, the TREE1-mediated transcriptional repression was not detected (Fig. 4 H–J and SI Appendix, Fig. S4C). All together, these data suggest that TREE1 is a transcriptional repressor, and EIN3 enhances TREE1 mediated transcriptional repression in plant cells.
为了探究TREE1与TREE1结合基序的结合活性,我们首先进行了EMSA实验。我们发现TREE1与TREE1结合基序存在强烈结合活性,另外TREE1与DRY2启动子含有TREE1结合基序的区域也存在强烈结合活性,而当基序被突变后就不再能检测到有结合活性(Fig. 4 A and B, Fig. S4A)。随后我们就将此基序命名为TREE1结合基序。接下来我们继续探究TREE1在转录调控中的功能。我们在农杆菌中构建了带有35s启动子驱动的萤光素酶以及5个EIN3结合基序和6个TREE1结合基序,将其在烟草中瞬时表达,以及另外包括GUS/EIN3/TREE1的表达(Fig. 4 C and D, Fig. S4A)。与GUS蛋白存在的对照组进行比较,EIN3存在时,萤光素酶信号显著增强。相反地,TREE1存在时,萤光素酶信号显著降低。更有趣的是,当TREE1与EIN3同时存在时,萤光素酶信号甚至更低(Fig. 4 E–G and SI Appendix, Fig. S4B)。当TREE1结合基序突变后,TREE1介导的转录抑制现象就不再存在了(Fig. 4 H–J and SI Appendix, Fig. S4C)。总而言之,这些数据表明TREE1是一种转录抑制子,EIN3会增强TREE1介导的转录抑制活动。
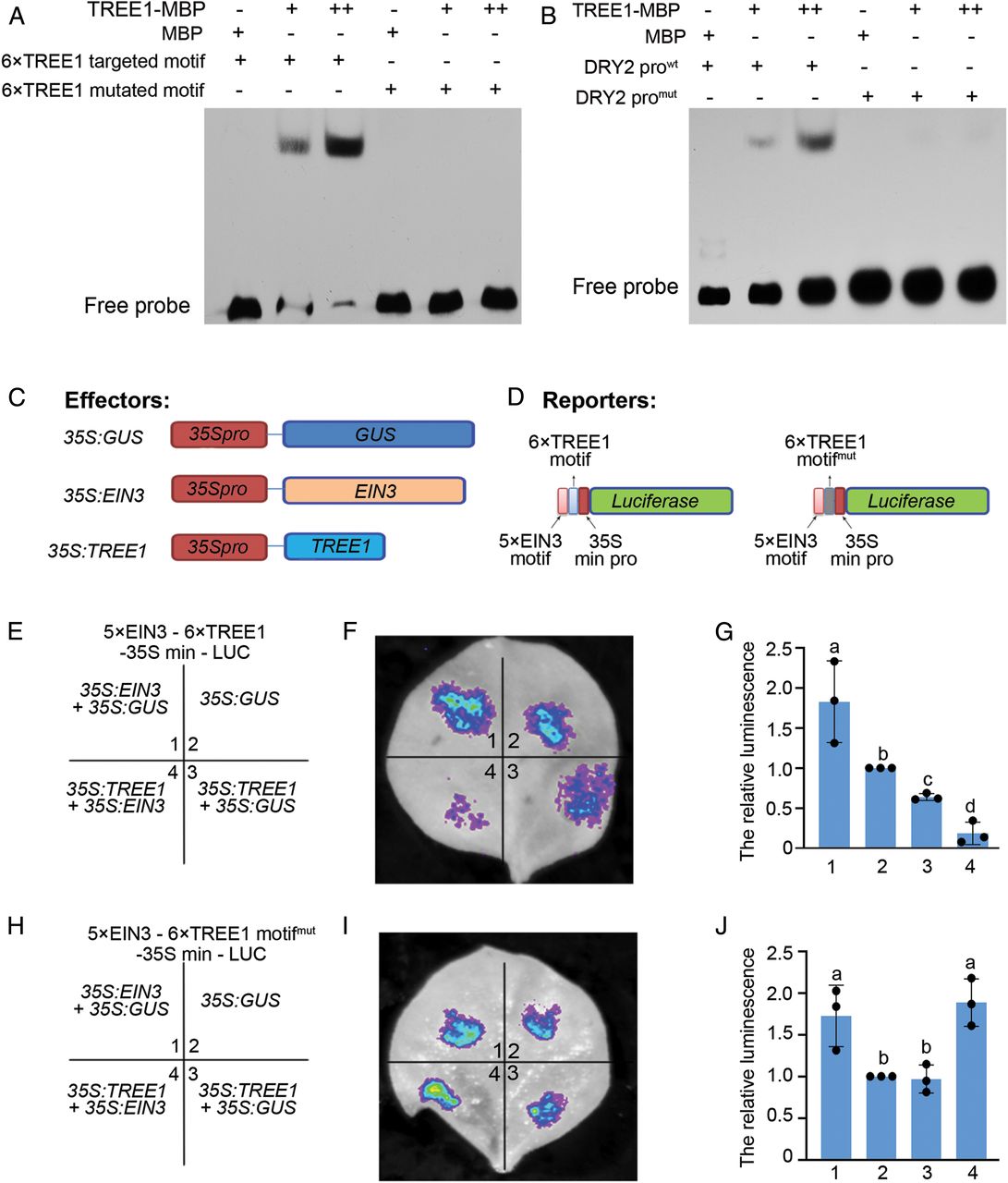
5.TREE1 Mediates Transcriptional Repression in Response to Ethylene to Inhibit Shoot Growth.
5.TREE1介导的乙烯响应下的转录抑制导致地上部分生长的阻滞
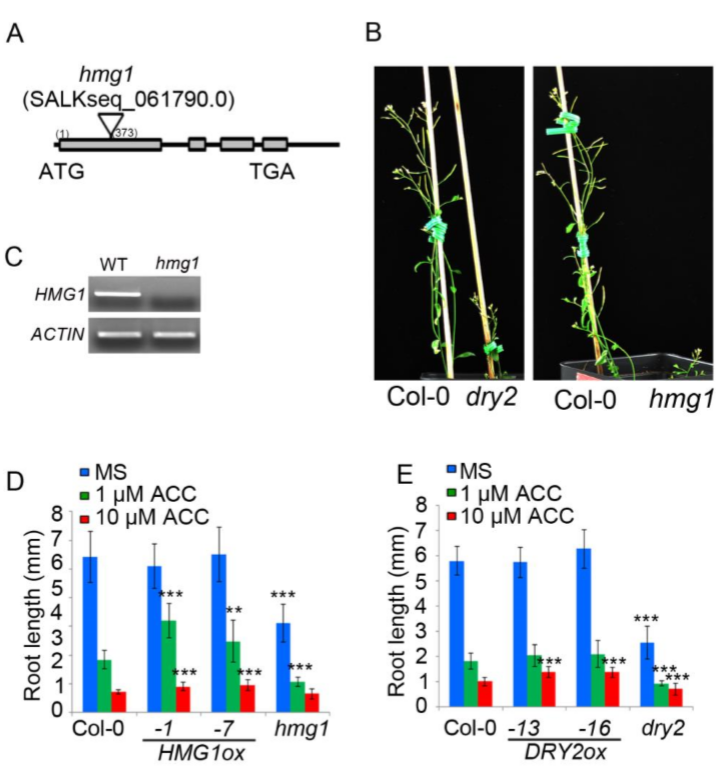
To further validate the function of TREE1 in the ethylene response, we first examined the TREE1 gene expression by qRT-PCR. We found that TREE1 mRNA levels were elevated by ethylene in shoots, but not in roots (SI Appendix, Fig. S5A). We then obtained TREE1 T-DNA mutants (tree1-1 and tree1-2)(SI Appendix, Fig. S5B); these plants did not display an obvious ethylene-responsive phenotype (SI Appendix, Fig. S5C). Given DAZ3 is the closest homolog of TREE1 and it locates on the same chromosome, we therefore mutated DAZ3 in the Col-0 or in the tree1-1 mutant using CRISPR-Cas9 gene editing to generate daz3 single mutant and daz3/tree1-1 double mutant (36) (SI Appendix,Fig. S5D and E).Similar to the tree1-1 single mutant, the daz3 single mutant did not display an obvious ethylene-responsive phenotype (Fig. 5 A and B and SI Appendix, Fig. S5F). But, the daz3tree1-1 double mutant displayed an ethylene insensitive phenotype in shoots (Fig. 5 A and B and SI Appendix, Fig. S5F). The ethylene insensitive phenotype in shoots was further confirmed by knocking down DAZ3 (DAZ3RNAi) in the tree1-1 mutant (DAZ3RNAi tree1-1)( SI Appendix, Fig. S5 G–J).
为了进一步验证TREE1在乙烯响应下的功能,我们首先用qrt检验了TREE1的表达量。我们发现TREE1在shoot中的mRNA水平会受乙烯调控增加,但root中并不会(Fig. S5A)。随后我们获得了TREE1的T-DNA插入突变体(tree1-1 & tree1-2)(Fig. S5B);这些植株并没有表现出非常显著的乙烯响应差异表型(Fig. S5C)。DAZ3是TREE1最近的同源基因,并且它们在同一个染色体上,所以我们利用CRISPR-CAS9获得了daz3单突突变体以及daz3/tree1-1双突突变体[36](Fig. S5D and E)。与tree1-1单突一样,daz3单突没有表现出显著的乙烯响应差异表型(Fig. 5 A and B, Fig. S5F)。但是daz3/tree1-1双突在shoot中表现出乙烯迟钝表型(Fig. 5 A and B, Fig. S5F)。将DAZ3进行Ri干扰后的表型进一步确认了乙烯迟钝的表型(Fig. S5 G–J)。
博主注:ACC (氨基环丙烷羧酸)是乙烯的直接生物合成前体。
Next, we generated the TREE1 and the DAZ3 gain-of-function plants (TREE1ox and DAZ3ox). We found that in the absence of ethylene, the TREE1ox plants displayed a dwarfed phenotype (Fig. 5C and SI Appendix, Fig. S5K). In the presence of ethylene, the plants displayed a more severe ethylene-responsive phenotype (Fig. 5C). But, no obvious ethylene responsive phenotype was observed in the DAZ3ox plants (Fig. 5C and SI Appendix, Fig. S5 K), suggesting that TREE1 plays a dominant role in the process. To further confirm the function of TREE1 in the ethylene response at a molecular level, we conducted transcriptome analyses in shoots. Compared to the Col-0, the ethylene-induced repression of gene expression was largely eliminated in the daz3tree1-1 double mutant (Fig. 5D and SI Appendix, Fig. S5 L and Table S1). In the TREE1ox plants, however, most of gene expression was repressed, even in the absence of ethylene (Fig. 5E and SI Appendix, Table S1); near 50% of the genes that were repressed by ethylene in Col-0 were also repressed in the TREE1ox plants without ethylene treatment (Fig. 5F). The qRT-PCR analysis in SI Appendix, Fig. S5M further confirmed these results.
Next, we examined the TREE1 binding targets by ChIP-seq in the shoots of TREE1p:TREE1-GFP/Col-0 transgenic seedlings by using GFP antibody (SI Appendix, Table S1). Over 50% of the TREE1 regulated genes were found to be bound by TREE1, under either air or ethylene treatment (SI Appendix, Fig. S5N). Near 50% of ethylene-repressed genes in shoots were binding targets of TREE1 (SI Appendix, Fig. S5O). Notably, the binding signals of TREE1 were elevated in the presence of ethylene compared to that without ethylene treatment (Fig. 5G). The ChIP-qPCR assay in the shoots of TREE1ox with or without 4 h of ethylene treatment further confirmed the ChIP-seq result (SI Appendix, Fig. S5 P). Additionally, a DNA binding motif that is similar to the TREE1 binding motif (Fig. 1G) was found in the top 200 TREE1 binding peak regions (SI Appendix, Fig. S5Q), and the TREE1 binding to this motif was further confirmed by an EMSA assay (SI Appendix, Fig. S5R). Taken together, these results strongly suggest that TREE1 is involved in the ethylene response as a transcriptional repressor.
随后,我们获得了TREE1以及DAZ3的功能增益植株(gain-of-function plants),即TREE1ox及DAZ3ox。我们发现,当乙烯不存在时,TREE1ox表现出矮小表型(Fig. 5C and SI Appendix, Fig. S5K),当乙烯存在时,TREE1ox表现出更加严重的乙烯响应表型(Fig. 5C)。但是DAZ3ox植株中没有显著表型(Fig. 5C and SI Appendix, Fig. S5K),这表明TREE1起了主导作用。为了进一步在分子水平上验证TREE1的功能,我们进行了shoot中的转录组分析。与野生型相比,daz3/tree1-1双突植株中,观察到了大量的乙烯促使的基因抑制表达(Fig. 5D and SI Appendix, Fig. S5 L)。在TREE1ox植株中,大多数基因都被抑制表达了,甚至是乙烯不存在的时候(Fig. 5E and SI Appendix, Table S1)。野生型中受乙烯下调表达的基因中将近50%在TREE1ox植株中,没有乙烯处理时也下调了(Fig. 5F)。qrt分析进一步证明了这些结果(Fig. S5M)。
随后,我们使用了GFP抗体利用CHIP-seq检测了pTREE1:TREE1-GFP/Col-0的shoot中的TREE1结合基序。超过50%的TREE1调控基因与TREE1有结合,无论是空气或者乙烯处理(Fig. S5O)。TREE1的结合信号在乙烯存在时更加显著(Fig. 5G)。CHIP-qPCR分析进一步验证了以上结果(Fig. S5 P)。另外,我们还发现了一个与之前TREE1结合基序非常相似的基序,EMSA实验也证实了该基序与TREE1的结合(Fig. S5Q,R)。总而言之,这些结果强有力地证明TREE1作为转录抑制子参与了乙烯响应。
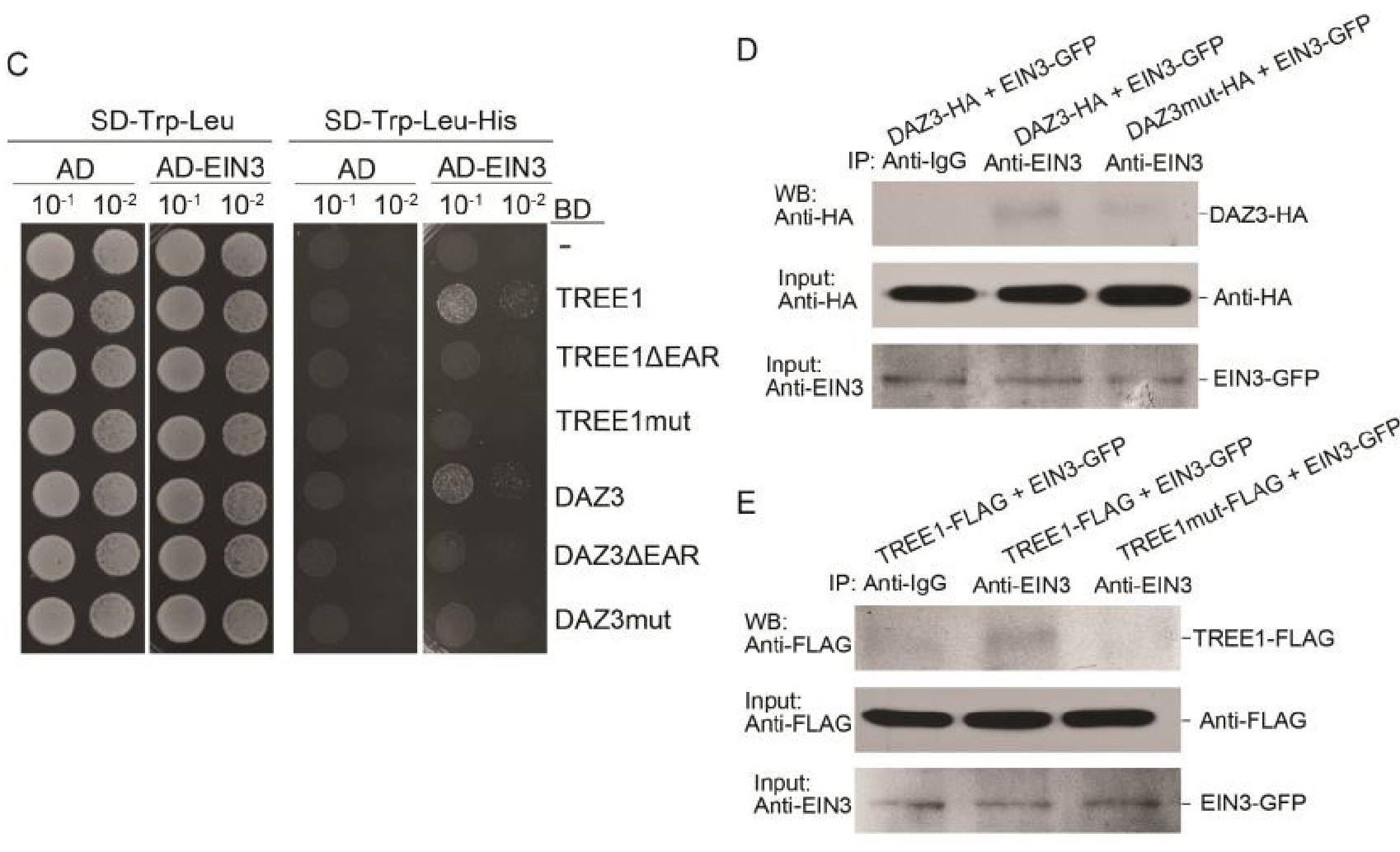
EAR motifs are known to mediate transcriptional repression (35, 37, 38); TREE1 has two of these motifs (SI Appendix, Fig. S3B), and the deletion/mutation of EAR motifs impaired the interaction between TREE1 and EIN3 (Fig. 3A and SI Appendix, Fig. S3 C–E), suggesting that EAR motifs play an important role in the ethylene-mediated transcriptional repression. To evaluate the function of EAR motifs in TREE1, we generated the plants that expressed TREE1 without EAR motifs (TREE1ΔEARox) (Fig. 5H and SI Appendix, Fig. S5S). Among all of the transgenic plants, no single TREE1ΔEARox line showed an obvious phenotype (Fig. 5I). A further qRT-PCR assay in shoots showed that the ethylene-mediated transcriptional repression in Col-0, or the enhanced ethylene-mediated transcriptional repression in the TREE1ox shoots, was almost undetectable in the TREE1ΔEARox shoots (Fig. 5J), demonstrating that EARs are important for the function of TREE1 in ethylene-mediated transcriptional repression.
EAR基序已经被证实会介导植物的转录抑制[35,37,38];TREE1含有两个EAR基序,并且EAR基序的突变或缺失会减弱TREE1与EIN3的互作,这表明EAR基序在乙烯介导的转录抑制中扮演了重要角色。为了验证TREE1中EAR基序的功能,我们获得了没有EAR基序的TREE1转基因植株(即TREE1ΔEARox)(Fig. 5H and SI Appendix, Fig. S5S)。shoot中的qrt分析显示,曾经在野生型中观察到的乙烯响应转录抑制,以及TREE1ox中shoot的更强烈的乙烯响应转录抑制,在TREE1ΔEARox shoots中并没有被观察到(Fig. 5J),这表明EAR基序对于TREE1调控乙烯响应下的转录抑制非常重要。
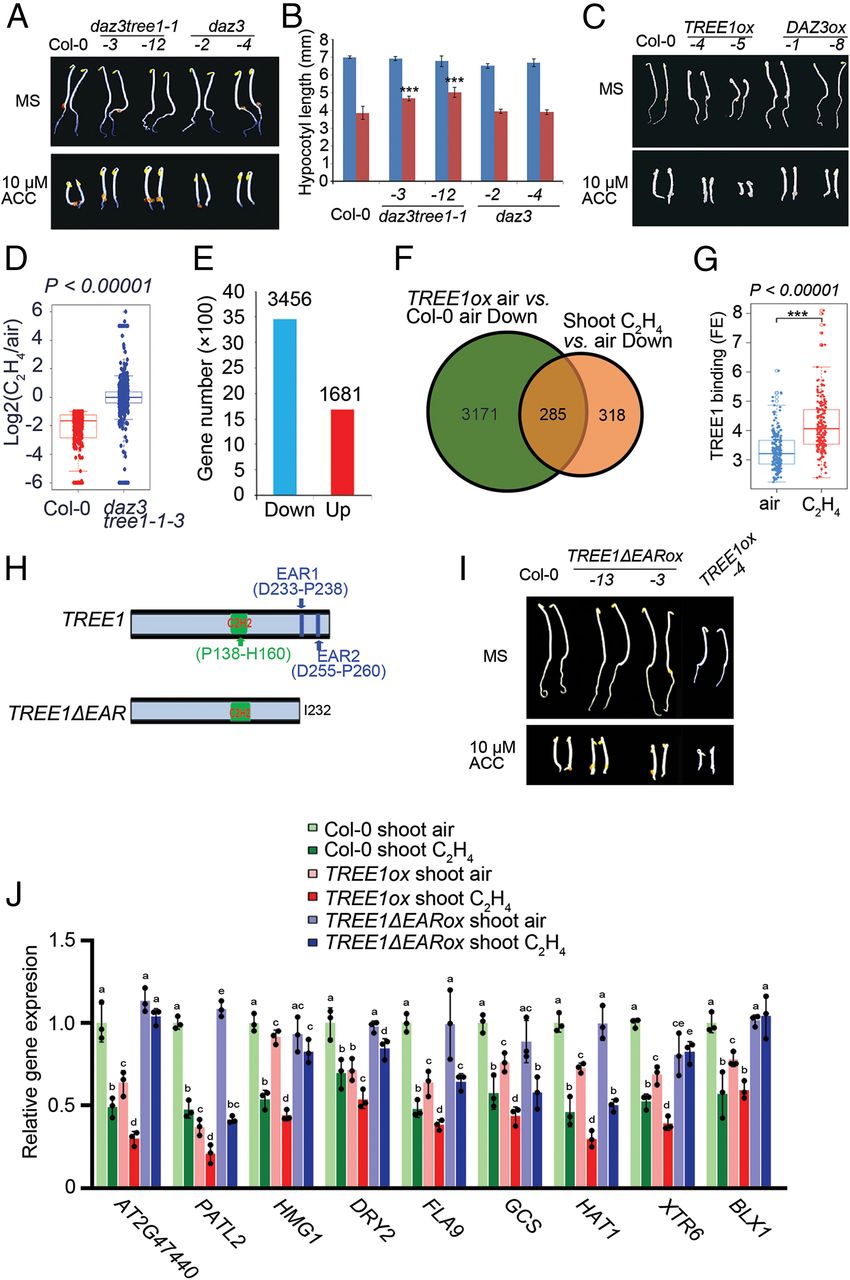
6.EIN3 Is Required for TREE1-Mediated Transcriptional Repression in Response to Ethylene.
6.EIN3在TREE1介导的乙烯调控下的转录抑制中具有必要性
To further explore the connection between TREE1 and EIN3, we first crossed TREE1ox and ein31eil1-1 plants to generate TREE1ox/ein3-1eil1-1 plants. The hyperethylene-sensitive phenotype in the shoots of TREE1ox was recovered in the TREE1ox/ein3-1eil1-1 (Fig. 6A and SI Appendix, Fig. S6 A and B). However, the TREE1 protein levels were not regulated by EIN3 and EIL1 (Fig. 6B). We next examined the expression of TREE1 binding genes in the shoots from both the ein3-1eil1-1 mutant and the EIN3 gain-of-function (EIN3ox) plants. The ethylene-mediated transcriptional repression detected in Col-0 shoots was not detectable in ein3-1eil1-1 shoots, whereas it was significantly enhanced in EIN3ox shoots (Fig.6C). In addition, ethylene-mediated transcriptional repression in the target genes was not detected in TREE1ox/ein3-1eil1-1 shoots (Fig. 6C). These results indicate that EIN3 is involved in the TREE1-mediated transcriptional repression at a molecular level.
为了进一步探索TREE1与EIN3之间的联系,我们首先获得了TREE1ox/ein3-1eil1-1植株。曾经在REE1ox植株中观察到的乙烯高度敏感表型被回复了(Fig. 6A and SI Appendix, Fig. S6 A and B)。然而,TREE1的蛋白表达水平并没有显著改变(Fig. 6B)。我们随后检测了在ein3-1eil1-1双突与EIN3ox植株中TREE1结合基因的表达量。之前野生型中乙烯下调表达的基因并没有被观察到,反而在EIN3ox中更显著下调了(Fig.6C)。另外,在TREE1ox/ein3-1eil1-1中也没有观察到显著的下调。这表明,EIN3参与了TREE1介导的转录抑制。
Next, we examined how EIN3 influences TREE1 binding by ChIP-qPCR in the shoots of TREE1ox and TREE1ox/ein3-1eil1-1 with or without 4 h of ethylene treatment (SI Appendix, Fig. S6 C–H). Compared to without the ethylene treatment, the TREE1 binding activity was elevated by ethylene treatment in TREE1ox shoots (Fig. 6D). The binding activity was reduced in TREE1ox/ein3-1eil1-1 shoots compared to that in Col-0 shoots, and the ethylene-induced elevation of TREE1 binding that was detected in TREE1ox shoots was abolished in TREE1ox/ein31eil1-1 shoots (Fig. 6D), showing that EIN3 enhances TREE1 binding, specifically in the presence of ethylene. Given that TREE1 binding is enhanced by ethylene treatment when EIN3 protein is accumulated (Fig. 5G), we decided to examine the influence of EIN3 on TREE1 binding by an EMSA assay. We found that the TREE1 binding activity was significantly enhanced by EIN3 and the activity was correlated with the amount of EIN3 proteins (SI Appendix, Fig. S6I), further confirming a positive regulation of EIN3 on the TREE1 binding activity. Together, all these results indicate that EIN3 is required for the TREE1-mediated transcriptional repression.
随后,我们利用CHIP-qPCR检测了在TREE1ox以及TREE1ox/ein3-1eil1-1中EIN3如何影响TREE1的结合(Fig. S6 C–H)。在TREE1ox shoot中,乙烯处理使TREE1的结合活性提升(Fig. 6D)。TREE1ox/ein3-1eil1-1植株中TREE1的结合活性降低(Fig. 6D)。这表明EIN3增强了TREE1的结合,尤其是当乙烯存在的时候。前文已经提到当乙烯处理时,EIN3蛋白的积累会使TREE1的结合活性增强(Fig. 5G),我们决定用EMSA实验验证EIN3对于TREE1结合活性的影响。结果显示,EIN3会让TREE的结合活性显著增强,并且结合活性还与EIN3蛋白的含量有关联(Fig. S6I),这进一步验证了EIN3对于TREE1结合活性的积极作用。总而言之,这些结果都显示,EIN3在TREE1介导的转录抑制中具有必要性。
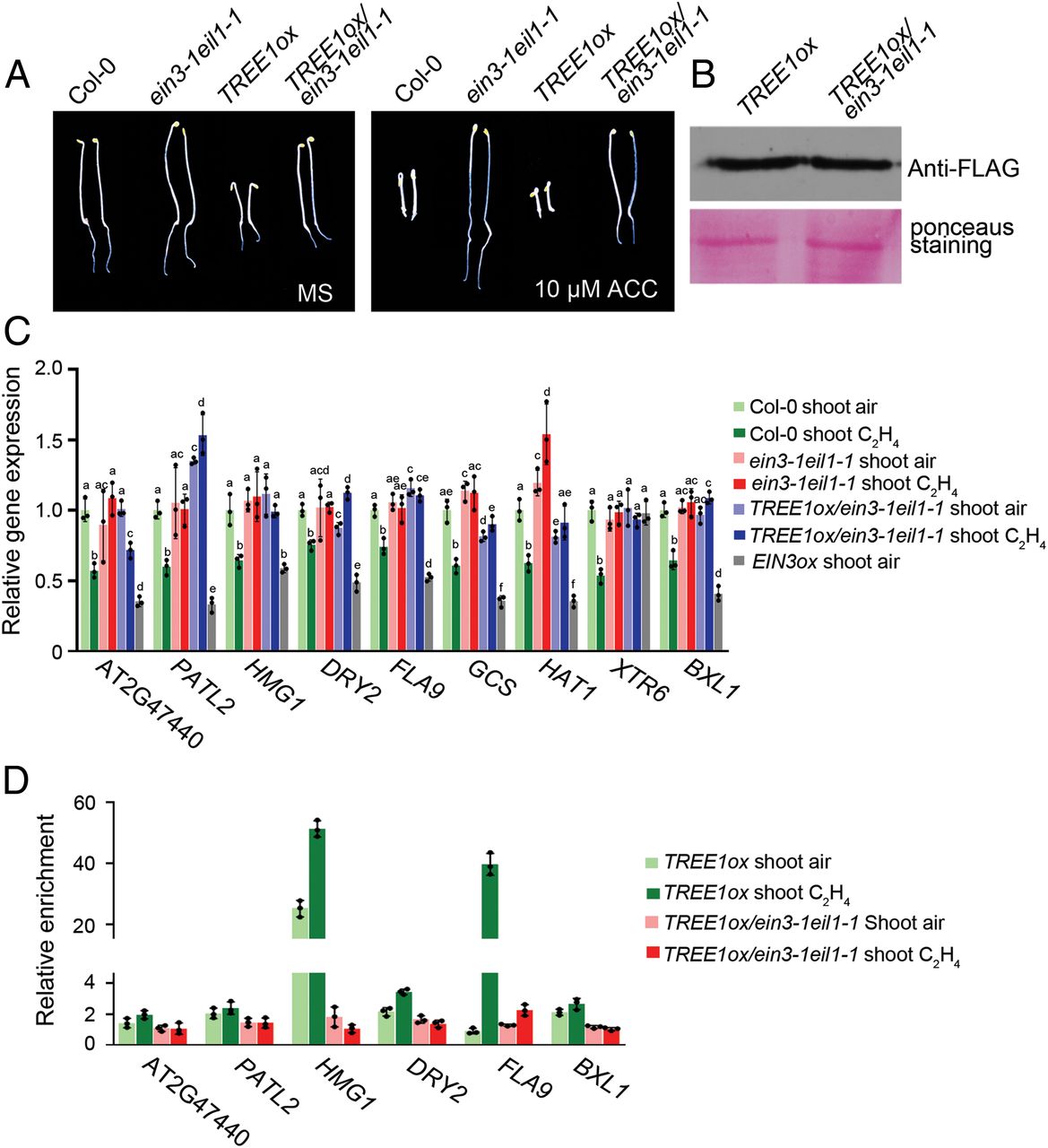
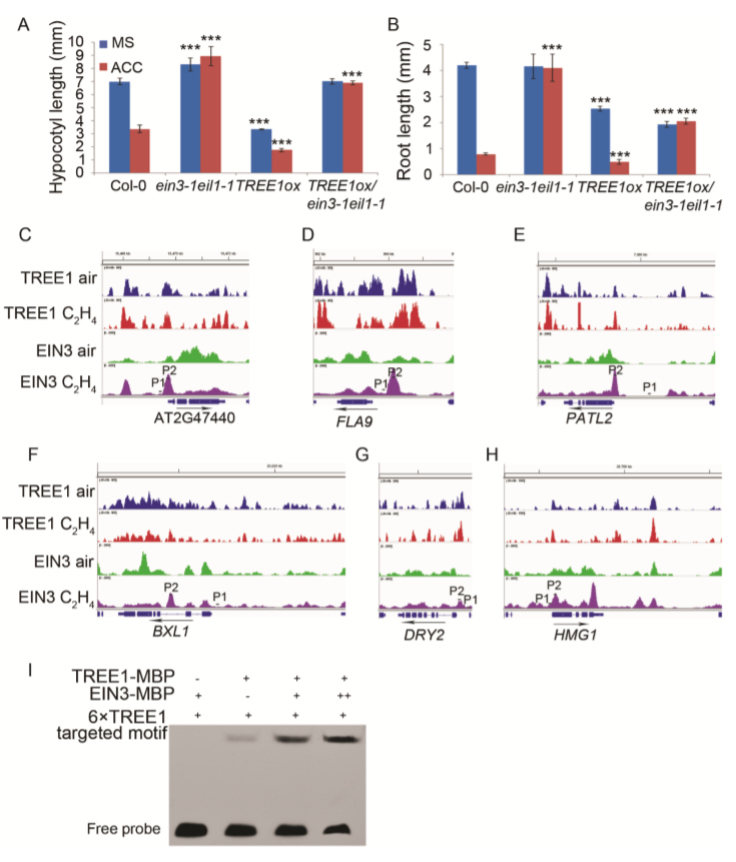
7.Plants with Mutations in Two of the TREE1 Targets Have Dwarfed Hypocotyl Phenotypes.
7.两种TREE1靶向基因的突变体植株都具有矮化下胚轴的表型特征
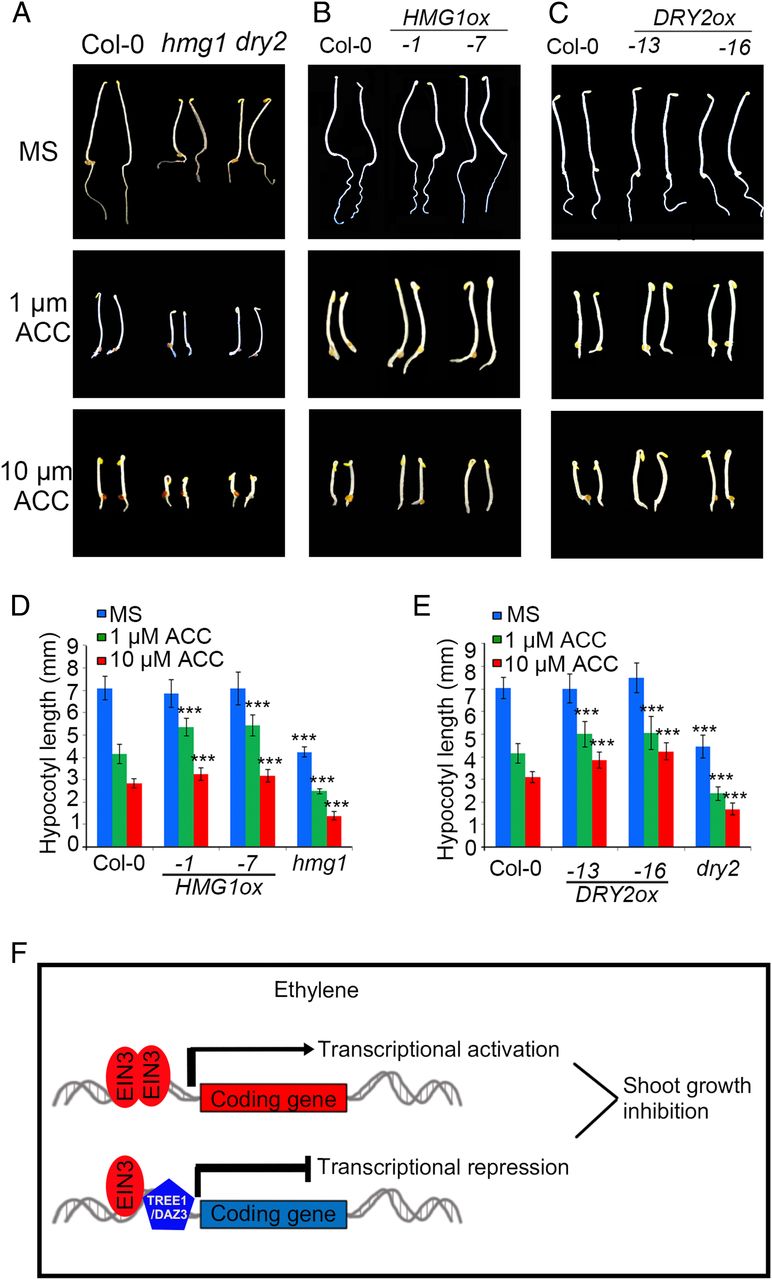
We hypothesized that in the presence of ethylene, the complex of TREE1-EIN3 represses gene expression, leading to an inhibition of shoot growth. To test our hypothesis, we obtained T-DNA insertion mutants for two TREE1 binding targets that are also bound by EIN3 and their gene expression was repressed by ethylene. Plants with mutations in either of these genes, HMG1 (HYDROXY METHYLGLUTARYL COA REDUCTASE 1) and DRY2, had similar dwarfed hypocotyl phenotypes in the absence of ethylene (Fig. 7A and SI Appendix, Figs. S2B and C and S7 A–C). We then generated their gain-of-function plants, and we found that the HMG1ox and DRY2ox plants were less sensitive to ethylene in shoots than that of Col-0 plants (Fig. 7 B–E and SI Appendix, Fig. S7D and E). Most interestingly, the adult plants were dwarfed, and they had a serious fertility problem (SI Appendix, Fig. S7 B). Taken together these data provide additional genetic evidence that TREE1 mediates transcriptional repression during the ethylene response to inhibit shoot growth.
我们推断,当乙烯存在时,TREE1-EIN3复合体调控了基因表达的抑制,导致shoot生长的停滞。为了验证我们的推断,我们选取了两个TREE1的结合靶向基因(它们同时会与EIN3结合),获得了这两个基因的T-DNA插入突变体,最终发现它们的表达量在乙烯调控下下调。这两个基因分别是HMG1 (HYDROXY METHYLGLUTARYL COA REDUCTASE 1) 和DRY2。无论是哪个基因的突变体植株,在乙烯不存在时都具有相似的表型——矮化的下胚轴(dwarfed hypocotyl)(Fig. 7A and SI Appendix, Figs. S2B and C and S7 A–C)。随后我们获得了它们的过表达植株,发现它们都在shoot中对乙烯敏感度降低(Fig. 7 B–E and SI Appendix, Fig. S7D and E)。更有趣的是,它们的成熟植株都是矮化的,并且它们都有严重的生殖问题(Fig. S7 B)。总而言之,这些数据都证明了TREE1介导的乙烯响应下的转录抑制活动,会导致地上部分的生长停滞,其中EIN3也非常重要。
Fig. 7 F: Model for the TREE1-mediated transcriptional repression in shoots. In the presence of ethylene, EIN3 alone targets the promoters of a subset of genes to activate transcription. In the promoters of a different subset of genes, the transcriptional repressor TREE1/DAZ3 interacts with EIN3 and transcription is repressed.
图:7F地上部分TREE1介导的转录抑制活动的机制模型。在乙烯存在时,EIN3会独自靶标一些基因去激活转录;但对于另一些基因,EIN3会与TREE1/DAZ3互作来抑制转录,这会共同导致植物地上部分生长发育的停滞。
[THE END]
文章pdf
2020-PNAS-TREE1-EIN3–mediated transcriptional repression inhibits shoot growth .pdf
文章supplementary materials
2020-PNAS-TREE1-EIN3–mediated-supp.pdf
供大家一起学习交流~
https://blog.sciencenet.cn/blog-3449010-1257876.html
上一篇:[Science]胚与胚乳之间的分子双向交流在种子发育过程中必不可少(3/END)
下一篇:[Mediat Inflamm, 2018] STING信号促进细胞凋亡、坏死以及细胞死亡:一篇综述及更新